Saiu no PRE de setembro...
Module identification in bipartite and directed networks
Roger Guimerà, Marta Sales-Pardo, and Luís A. Nunes Amaral -->
Northwestern Institute on Complex Systems (NICO) and Department of Chemical and Biological Engineering, Northwestern University, Evanston, Illinois 60208, USA
(Received 7 June 2007; published 6 September 2007)
Modularity is one of the most prominent properties of real-world complex networks. Here, we address the issue of module identification in two important classes of networks: bipartite networks and directed unipartite networks. Nodes in bipartite networks are divided into two nonoverlapping sets, and the links must have one end node from each set. Directed unipartite networks only have one type of node, but links have an origin and an end. We show that directed unipartite networks can be conveniently represented as bipartite networks for module identification purposes. We report on an approach especially suited for module detection in bipartite networks, and we define a set of random networks that enable us to validate the approach.
©2007 The American Physical Society
URL: http://link.aps.org/abstract/PRE/v76/e036102
doi:10.1103/PhysRevE.76.036102
PACS: 89.75.Hc, 89.65.-s, 05.50.+q
Additional Information
Full Text: PDF GZipped PS
Unveiling community structures in weighted networks
Nelson A. Alves -->
Departamento de Física e Matemática, FFCLRP Universidade de São Paulo, Avenida Bandeirantes 3900, CEP 14040-901, Ribeirão Preto, São Paulo, Brazil
(Received 28 February 2007; published 4 September 2007)
Random walks on simple graphs in connection with electrical resistor networks lead to the definition of Markov chains with transition probability matrix in terms of electrical conductances. We extend this definition to an effective transition matrix Pij to account for the probability of going from vertex i to any vertex j of the original connected graph G. Also, we present an algorithm based on the definition of this effective transition matrix among vertices in the network to extract a topological feature related to the manner by which graph G has been organized. This topological feature corresponds to the communities in the graph.
©2007 The American Physical Society
URL: http://link.aps.org/abstract/PRE/v76/e036101
doi:10.1103/PhysRevE.76.036101
PACS: 89.75.Hc, 05.10.-a
Additional Information
Full Text: PDF GZipped PS
Figura:
The protein folding network
MD simulations of aggregation
F. Rao and A. Caflisch, J. Mol. Biol. 342, 299-306 (2004) [pdf] [Supp. Mat.]
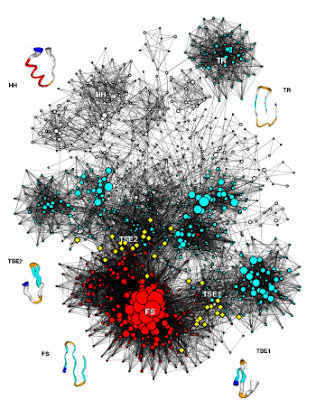
The protein folding network
MD simulations of aggregation
F. Rao and A. Caflisch, J. Mol. Biol. 342, 299-306 (2004) [pdf] [Supp. Mat.]
Beta3s conformation space network. The size and color coding of the nodes reflect the statistical weight and average neighbor connectivity, respectively. Representative conformations are shown by a pipe colored according to secondary structure: white stands for coil, red for alpha-helix, orange for bend, cyan for strand and the N-terminus is in blue. The variable radius of the pipe reflects structural variability within snapshots in a conformation. The yellow diamonds are folding TS conformations
Comentários